DX Stories: How We’re Making Whole Genome Sequencing in Patient Care a Reality

Source: https://blog.westerndigital.com/whole-genome-sequencing-in-patient-care/
Author: Wim De Wispelaere
All of us who are entrepreneurs see a better way to do something, or a better way to solve a problem. When we initially architected ActiveScale™, we knew that data is growing at incredible rates and that petascale solutions are going to be key to unlocking the possibilities of data.
We didn’t know all of the potential use cases for what we were creating back then, nor how many of them will be about advancing mankind or have a direct impact on our lives. I’m excited to share with you one such recent use case that’s bringing next generation whole genome sequencing in patient care one step closer to reality.
The Hurdle to Clinical Genomics is Not Just $$$
Since the first human genome was published, the scientific community agreed that if we were able to achieve affordable genome sequencing technologies, we’d have the potential to revolutionise patient care and medical treatment.
Today, with the introduction of next-generation sequencing, a whole human genome can be sequenced in a matter of days and at a cost that has dropped down from millions of dollars to below $1,000 dollars.
These new machines are quickly eliminating the time and cost hurdle of sequencings for clinical use.
Yet whole genome analysis and its interpretation – turning the raw sequence of over 3 billion base pairs in each individual’s genome into ‘knowledge’ and useful information for doctors – remains a challenging endeavour. Analysis can still take several days to weeks, even when using state of the art algorithms and compute infrastructure.
Why Whole Genome Sequencing?
Due to cost and time challenges, sequencing is often limited to the protein-coding regions of the genome, referred to as the exome. The reason is that the exons that compose the exome is where most known mutations that cause disease occur. Yet the exome represents only one percent of the whole genome …
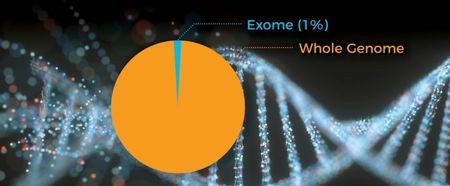
Researchers have found that variants in other regions of the genome can equally lead to severe health conditions. Hence, for medical practitioners, the analysis of the exome is as limiting as looking through a keyhole, or evaluating a patient by looking at just one limb. The ability to do whole genome analysis will throw the door wide open to expose the full spectrum of genome variants in which they could find indicators of even the rarest diseases.
The Genome Analytics Platform (GAP) Project
We’ve been involved in a very exciting research collaboration project in Belgium for the past two years, thanks to significant funding from the Flemish regional government. We teamed up with imec (one of the world’s leading R&D and innovation hub in nanoelectronics and digital technologies), the academic hospital UZ Leuven, the university KU Leuven, Ghent University, Agilent, and BlueBee to develop a unique hybrid cloud platform that can tackle this genome analytics challenge.
The goal was to build a platform that can make whole genome sequencing and analysis be both cost- and time-effective so hospitals can use it in day to day patient care. This was not simply a research project, but a specific real-world implementation request for the treatment of patients at UZ Leuven.
The Goal: to Analyze 48 Genomes, in Less Than 48 Hours
UZ Leuven is one of Europe’s leading medical research centres. Their goal was to move their genome research from focusing on the exome to the entire genome, and to make it a tool for daily patient care.
The hospital recently acquired a next-generation sequencer that is capable of sequencing 48 whole human genomes in just 48 hours. The target for the GAP project was to perform the data analysis and identify the genome variants in all 48 genomes in the same timeframe (less than 48 hours). In this manner, the hospital could run the sequencer in a continuous and most cost-efficient operation while doctors would be able to take quick medical decisions based on the genomic analysis, particularly for rare cases, rare diseases and complex disorders of new-borns.
This set us quite a challenge. The best practice GATK (genome analysis toolkit) from the Broad Institute takes 12 days per genome analysis in a single threaded deployment, or five days per genome on a 24-core machine. This required a new way of thinking about storage and compute infrastructure.
Building the Genome Analytics Platform (GAP)
There were many considerations in building the Genome Analytics Platform (GAP):
1. Dealing with massive data – First we had to ensure a dense storage solution that can deliver the throughput, data durability and capacity scaling capabilities required to accomplish imec’s extreme data-driven GAP goals. The ActiveScale object storage system was the obvious candidate for the task, as it can scale up to 63PB (raw) in a single namespace, deliver up to 19 nines of data durability, and offers unified data access (UDA). UDA allows both file-based and object-based data to be used natively on the system.
In this specific environment, the UDA nfs interface was implemented as it required no changes to the existing software that is used for ingesting the massive amounts of data from the sequencer or for reading the data in the analytics compute cluster. Moreover, the UDA nfs interface proved to deliver the required throughput to support the performance goal.
2. Making sure data doesn’t slow down – Once the genome data is loaded in the compute cluster, the data analysis algorithms requires massive parallel and random access to the data to align the samples, map them to the reference genome and identify the variants. To achieve our performance goals, we implemented Ultrastar® NVMe™ SSDs in the compute nodes and leveraged their extreme low latency and high random read performance to sustain the velocity of data. In the future, there will be an opportunity to leverage emerging technologies such as SCM to even further reduce processing time.
3. Laser focus on the algorithms – Speeding up the process of analysis did not only depend on next-generation hardware design, it required rethinking the analysis process. While we tuned the storage and hardware resources, University researchers from IMEC and Ghent University heavily optimised GATK algorithms to reduce the time needed for completion, so that together we could meet our 48-hour goal. Those optimised tools, called Halvade and elPRep, are open source and available to the worldwide community to use.
4. To cloud or not to cloud? – Finally, we needed to think of how to build an optimized architecture to support various workloads. For our specific workload of storing and analysing 48 whole genomes every 48 hours, the on-site Active Scale cloud storage, with the specifically tuned compute cluster, provides the most cost-efficient solution. However, in day-to-day operation, the hospital will be processing a variety of exome, whole genome sequences and research samples – workloads that will require varying compute power and varying time constraints.
In addition, the analysis of rare diseases often requires correlating data with samples from other research institutes around the globe. To cope with the variability of workloads, as well as the need for cloud collaboration, we decided to create a flexible hybrid solution, that can combine the best of both worlds; cost-effective, fast and low latency analysis at a local site, and bursting capabilities involving cloud computing. The hybrid architecture also allows to archive all data locally, with an optional DR copy in the cloud.
We Did It!
Together, through this collaborative effort, we built a new platform that can perform a full genome analysis of 48 samples in only 48 hours and at an acceptable cost. This allowed UZ Leuven to accelerate their analysis process per genome from five days to just four hours and opened new opportunities for their patient care that were once thought impossible. These possibilities pave the way for genome sequencing as a daily practice in more hospitals around the world. And, I’m proud to share that it is one of several medical research initiatives Western Digital is supporting in the field of genomic analysis and precision medicine diagnoses at scale.
Learn more about what Western Digital is doing in healthcare and research.

_______________________________________________________________________________________________________________________________________
To discover more case studies, why not register FREE for Digital Transformation EXPO Manchester!
_______________________________________________________________________________________________________________________________________
